
Replicate Weights
Methods Meetup: Session 1
Eve Perry
September 5, 2023
Outline
- Complex sampling recap
- What are replicate weights?
- Creating our replicate weights
- Using replicate weights in analyses
Complex Sampling Recap
Sampling Designs
Simple Random
- Only 1 level
- Each observation has same chance of selection
- Sample is often very small relative to population
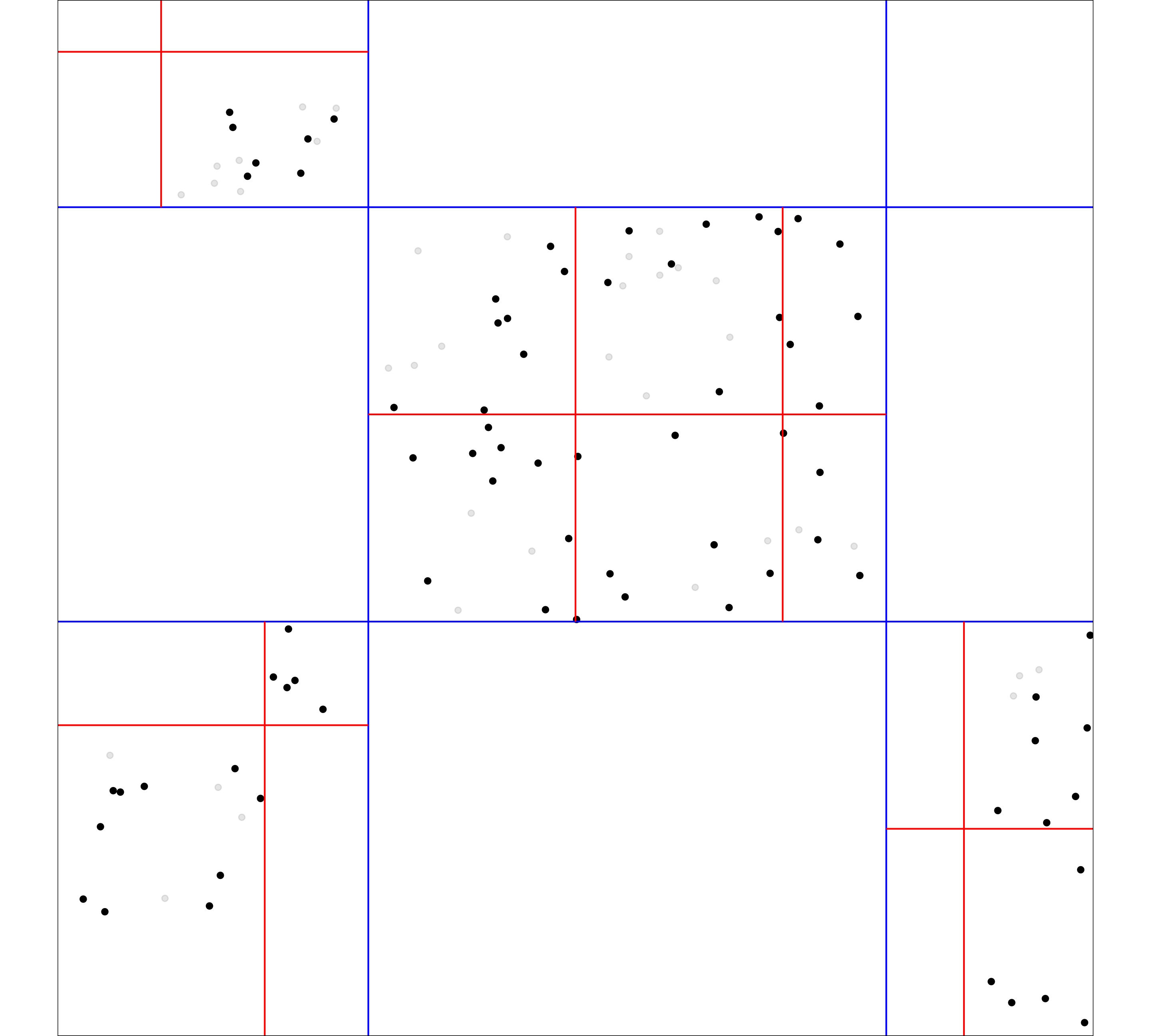
CASPEH
- 3 levels: county, venue, individual
- All sampled without replacement
- Counties and venues sampled with unequal probability
- Individuals sampled with equal probability
- Large fractions of populations sampled
Simple Random
CASPEH
Standard Error Formulas
Simple Random
\[ \sqrt{(1-\frac{n}{N})\frac{variance}{n}} \]
CASPEH

What Are Replicate Weights?
Concepts
- Compute standard errors when formulas are too complicated or inappropriate
- Use the sample as if it is itself a population
- Incorporate sampling design into the calculation of the weights
- Create multiple versions (replicates) of the sample and use variability in those versions for standard errors
- Come in a variety of forms: bootstrap, balanced repeated replication (BRR), jackknife
Bootstrap Resampling
- Draw a new sample from the original sample with replacement using the actual selection probabilities
- Follow the same sampling structure so it’s incorporated into the replicates
- Make an adjustment to the main weights so the replicate resembles the full sample
- Repeat 100 times
Creating Our Replicate Weights
Selection Probabilities
Counties & Venues
- Sampled without replacement and unequal probabilities
- Simulate the sampling procedure many (10,000-100,000) times to get the probability a given county and venue are selected
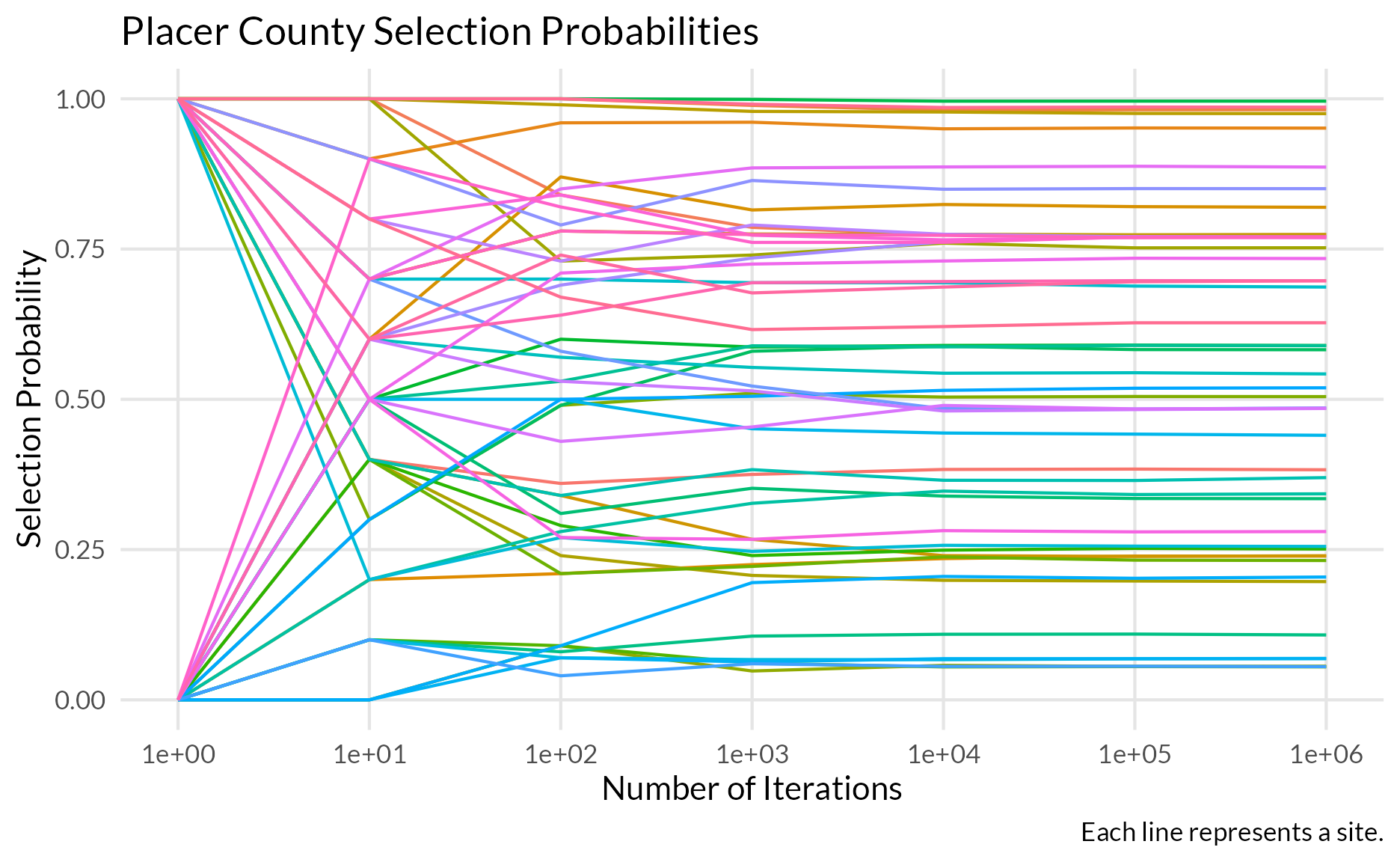
Individuals
- Simple random sample at venue
- Adjustments for individual non-response
- Adjustments for RDS inclusion
R: Packages
- Relies on
surveyandsvreppackagessurveyis the R workhorse for survey data analysissvrepis the only package which supports multi-stage, unequal probability sampling without replacement and large fractions of population
- Code available on the on-prem (git.ucsf.edu) GitHub
R: Code
design_object <- svydesign(
ids = ~ county + venue_2 + record_id,
probs = ~ county_probability + venue_probability + final_individual_probability,
fpc = ~ county_probability + venue_probability + final_individual_probability,
pps = "brewer",
data = design_data
)
options(list(survey.lonely.psu = "adjust", survey.replicates.mse = TRUE))
replicate_weights_design <- as_bootstrap_design(
design = design_object,
replicates = 100,
type = "Rao-Wu-Yue-Beaumont",
mse = TRUE,
samp_method_by_stage = c("PPSWOR", "PPSWOR", "SRSWOR")
)Using Our Replicate Weights
Conceptual Recap
- Estimate statistic of interest using main weights and each set of replicate weights
- Standard error comes from the variability of the replicate estimates with respect to the main weight estimate
R: Data Setup
R: Survey Setup
R: Chi-square Test
R: Regression
Call: svrepdesign.default(ids = ~record_id, weights = ~academic_weight,
repweights = "academic_rep_weight*", type = "bootstrap",
mse = TRUE, data = surveys)
Survey bootstrap with 100 replicates and MSE variances.
Call: svyglm(formula = episode_length ~ race_7cat, survey_object)
Coefficients:
(Intercept) race_7catNH White
40.366 9.158
race_7catNH Black race_7catNH AAPI
-2.819 16.096
race_7catNH Native American/Alaskan race_7catNH Multiracial
9.247 -2.846
race_7catNH Other
15.916
Degrees of Freedom: 3139 Total (i.e. Null); 93 Residual
(59 observations deleted due to missingness)
Null Deviance: 11750000
Residual Deviance: 11650000 AIC: 35910R: Group Means
library(srvyr)
survey_object %>%
as_survey() %>%
group_by(race_7cat) %>%
summarise(survey_mean(episode_length, na.rm = TRUE))# A tibble: 8 × 3
race_7cat coef `_se`
<fct> <dbl> <dbl>
1 Latinx/Hispanic/Indigenous 40.4 2.50
2 NH White 49.5 3.56
3 NH Black 37.5 3.03
4 NH AAPI 56.5 6.98
5 NH Native American/Alaskan 49.6 5.63
6 NH Multiracial 37.5 2.23
7 NH Other 56.3 15.3
8 <NA> 34.2 5.59Stata: Setup
use "${bhhi_shared_drive}/statewide_survey_processed_data/latest/statewide_survey_processed.dta"
keep if survey_count == 1
svyset _n [pweight=academic_weight], ///
bsrweight(academic_rep_weight*) ///
vce(bootstrap) ///
mse Sampling weights: academic_weight
VCE: bootstrap
MSE: on
Bootstrap weights: academic_rep_weight_1 .. academic_rep_weight_100
Single unit: missing
Strata 1: <one>
Sampling unit 1: <observations>
FPC 1: <zero>Stata: Regression
(running regress on estimation sample)
Bootstrap replications (100)
----+--- 1 ---+--- 2 ---+--- 3 ---+--- 4 ---+--- 5
.................................................. 50
.................................................. 100
Survey: Linear regression Number of obs = 3,140
Population size = 115,572.49
Replications = 100
Wald chi2(6) = 17.75
Prob > chi2 = 0.0069
R-squared = 0.0078
---------------------------------------------------------------------------------------------
| Observed Bstrap *
episode_length | coefficient std. err. z P>|z| [95% conf. interval]
----------------------------+----------------------------------------------------------------
race_7cat |
NH White | 9.157548 4.801016 1.91 0.056 -.2522703 18.56737
NH Black | -2.81859 4.02318 -0.70 0.484 -10.70388 5.066699
NH AAPI | 16.09569 7.197665 2.24 0.025 1.988528 30.20286
NH Native American/Alaskan | 9.246603 5.991962 1.54 0.123 -2.497426 20.99063
NH Multiracial | -2.846081 2.898077 -0.98 0.326 -8.526207 2.834045
NH Other | 15.91599 16.3178 0.98 0.329 -16.06631 47.89828
|
_cons | 40.36602 2.482923 16.26 0.000 35.49958 45.23246
---------------------------------------------------------------------------------------------Stata: Group Means
(running mean on estimation sample)
Bootstrap replications (100)
----+--- 1 ---+--- 2 ---+--- 3 ---+--- 4 ---+--- 5
.................................................. 50
.................................................. 100
Survey: Mean estimation Number of obs = 3,140
Population size = 115,572.49
Replications = 100
-----------------------------------------------------------------------------
| Observed Bstrap *
| mean std. err. [95% conf. interval]
----------------------------+------------------------------------------------
c.episode_length@race_7cat |
Latinx/Hispanic/Indigenous | 40.36602 2.482923 35.49958 45.23246
NH White | 49.52357 3.537419 42.59036 56.45678
NH Black | 37.54743 3.010734 31.6465 43.44836
NH AAPI | 56.46171 6.94129 42.85703 70.06639
NH Native American/Alaskan | 49.61262 5.600275 38.63629 60.58896
NH Multiracial | 37.51994 2.218442 33.17188 41.86801
NH Other | 56.28201 15.18007 26.52963 86.03439
-----------------------------------------------------------------------------Questions and Discussion
Additional Resources
- Stata Manual: Bootstrap Replicate Weights
- R
surveyPackage - IPUMS: Replicate Weights in the ACS
- Sharon Lohr: Sampling Design and Analysis
- R
svrepPackage - Beaumont and Emond (2022): A Bootstrap Variance Estimation Method for Multistage Sampling and Two-Phase Sampling When Poisson Sampling Is Used at the Second Phase
- Rao, Wu, Yue (1992): Some Recent Work on Resampling Methods for Complex Surveys

BHHI Methods Meetup Session 1: Replicate Weights